Breathing fresh air into respiratory research with single-cell RNA sequencing | European Respiratory Society

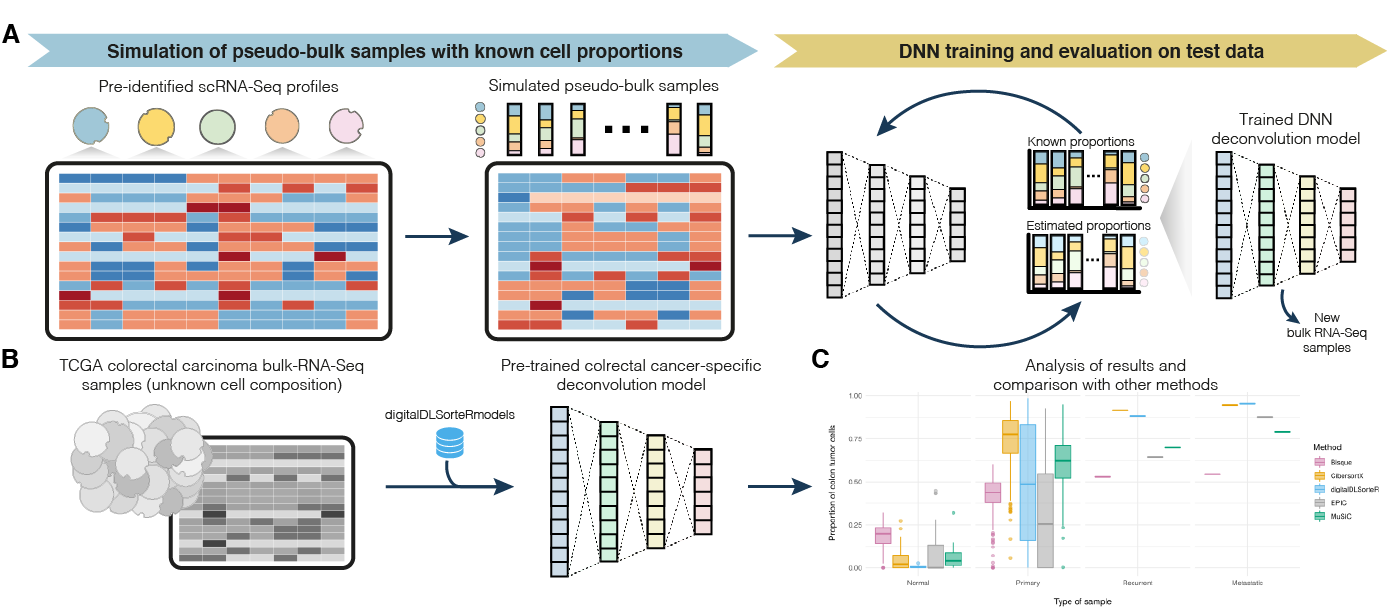
CDSeq – A novel complete deconvolution method for dissecting heterogeneous samples using gene expression data | RNA-Seq Blog

Single-cell RNA-sequencing analyses identify heterogeneity of CD8+ T cell subpopulations and novel therapy targets in melanoma: Molecular Therapy - Oncolytics

Integrative single-cell and bulk RNA-seq analysis in human retina identified cell type-specific composition and gene expression changes for age-related macular degeneration | bioRxiv

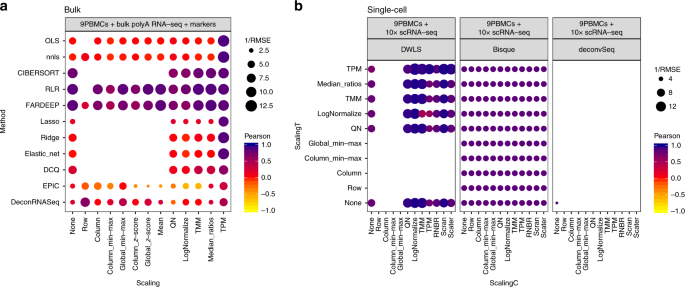
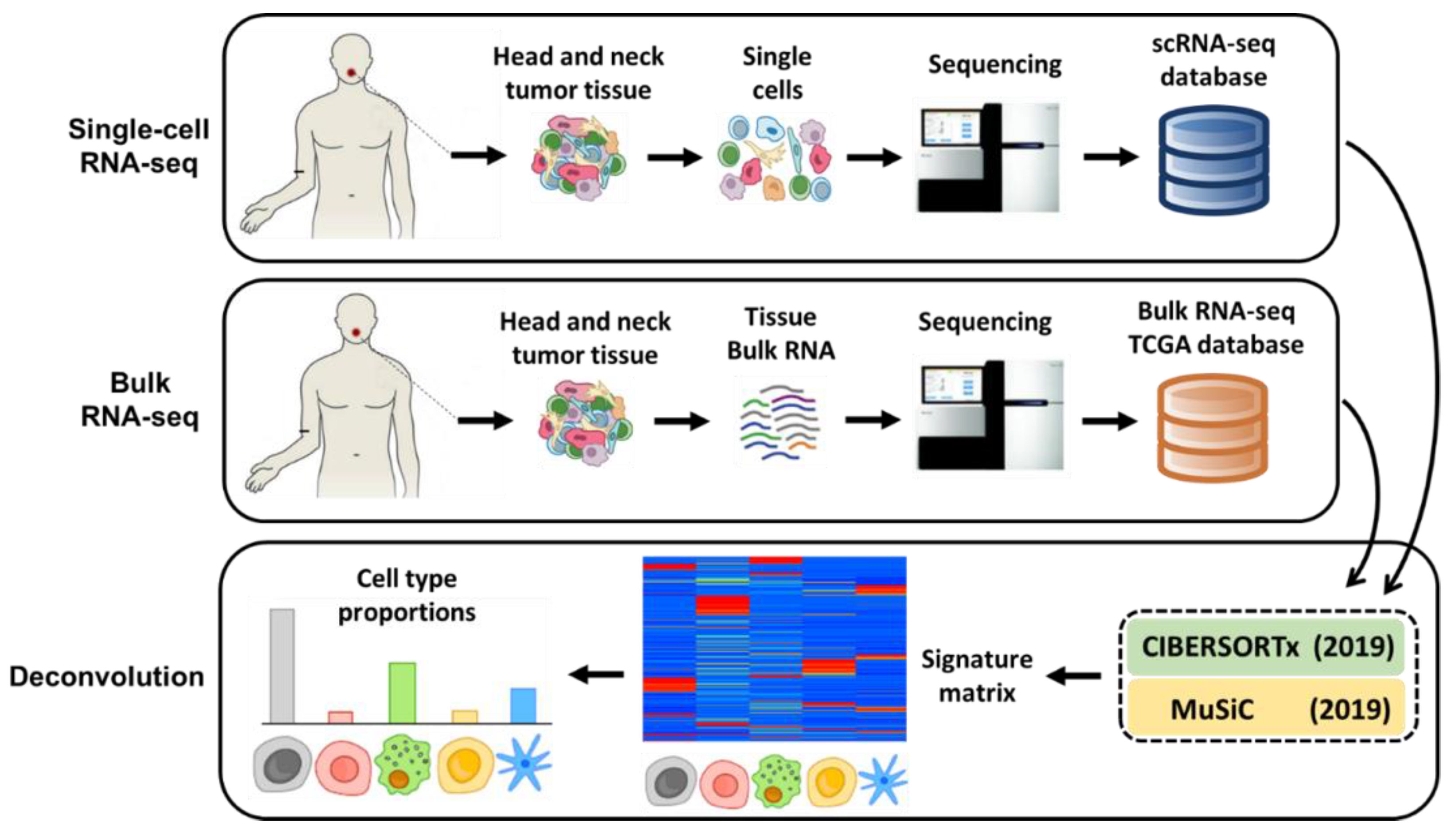
Illustration of popular methods used to analyse bulk and single-cell... | Download Scientific Diagram
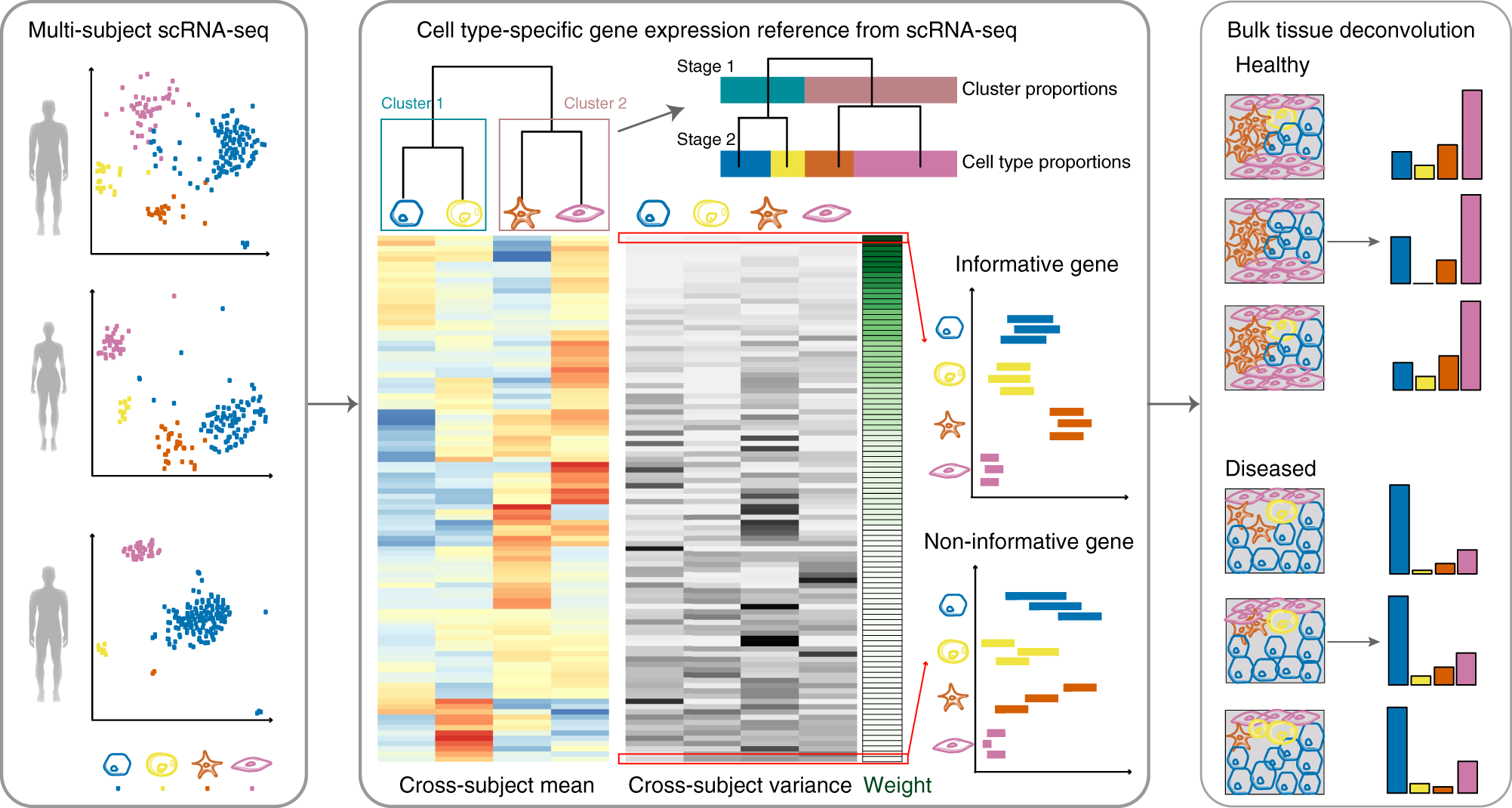
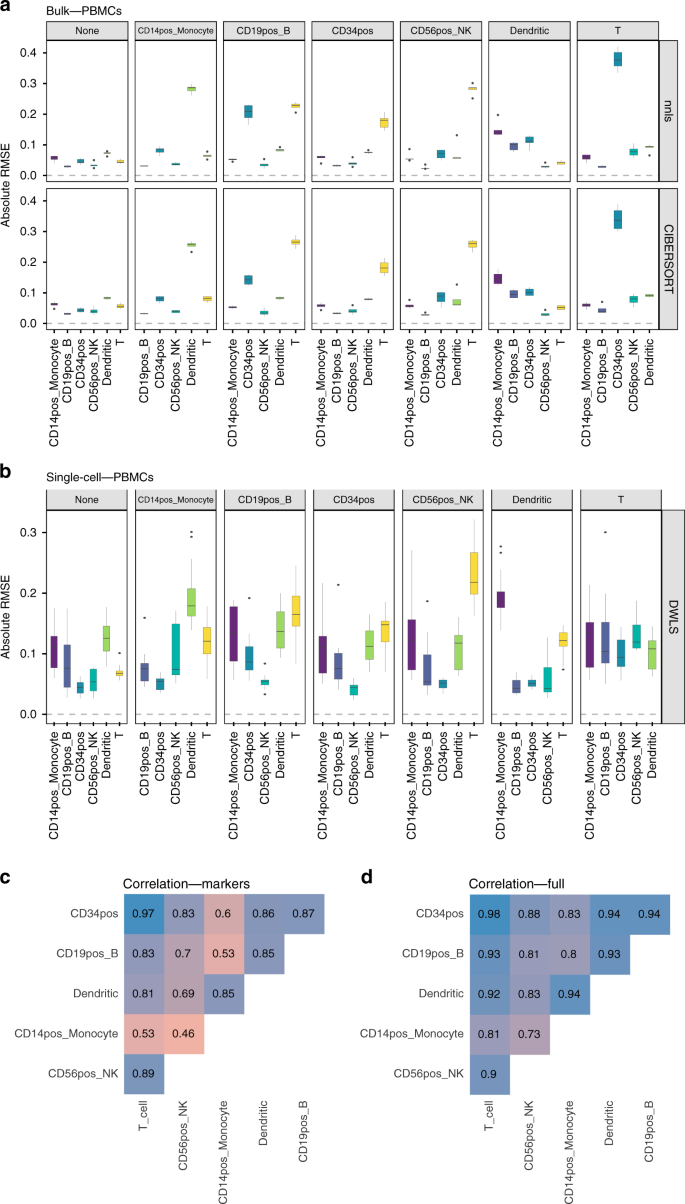
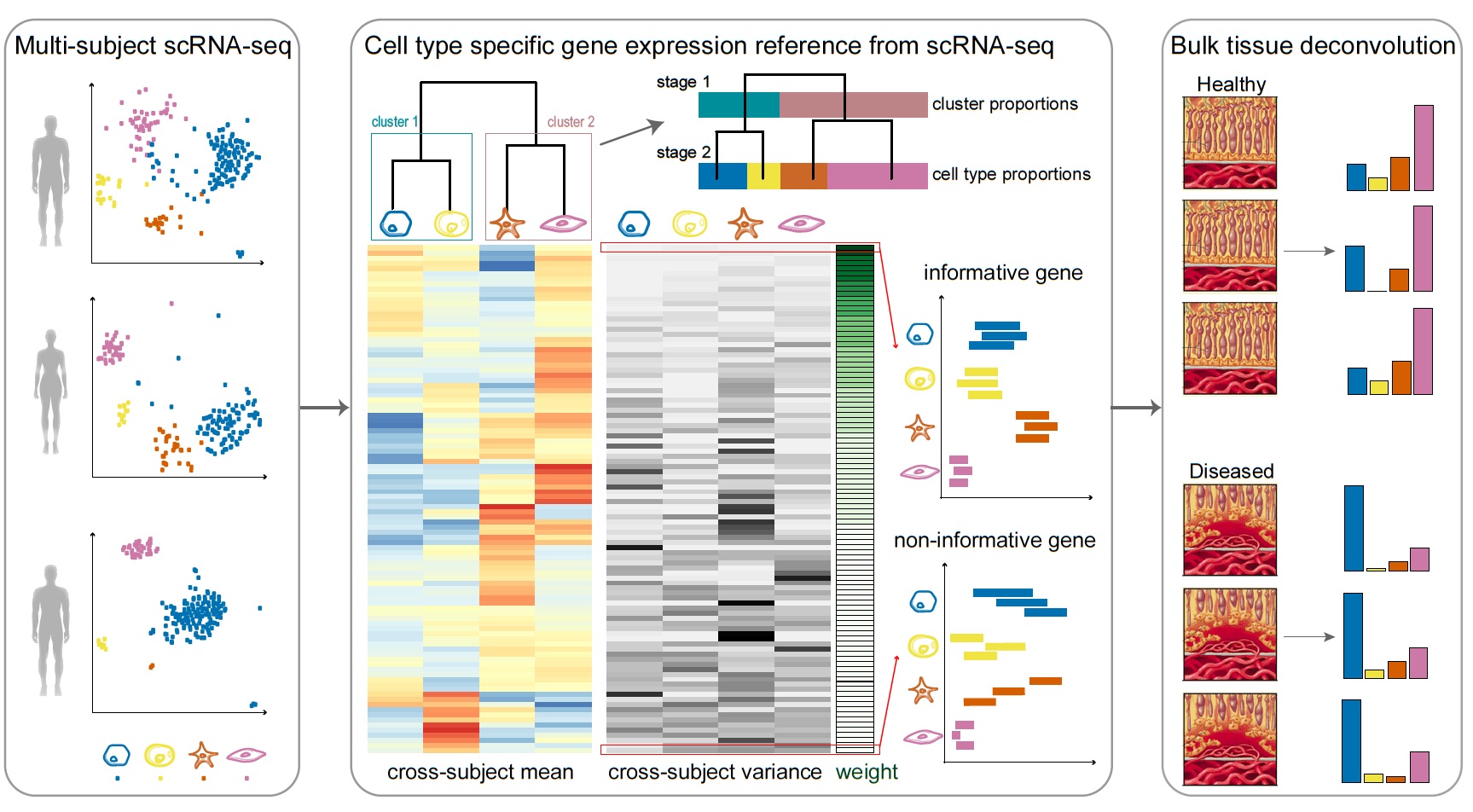
Bulk tissue cell type deconvolution with multi-subject single-cell expression reference | Nature Communications

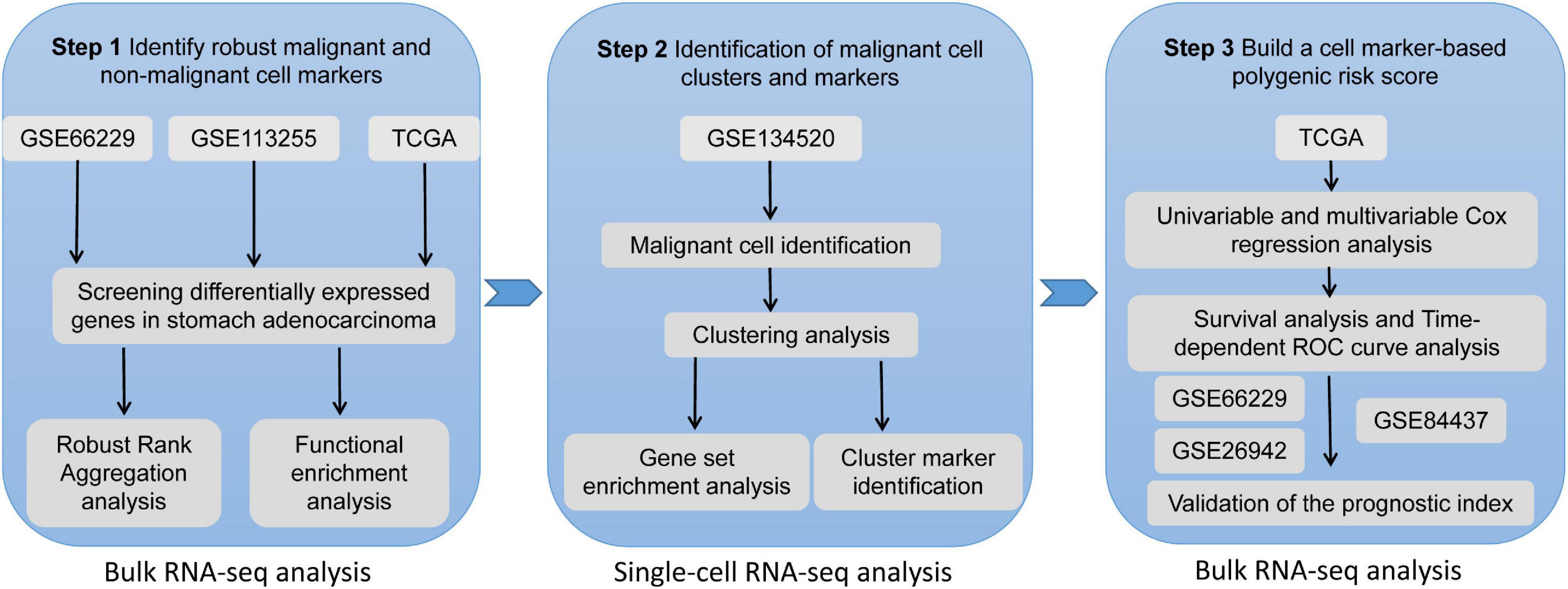
A protocol to extract cell-type-specific signatures from differentially expressed genes in bulk-tissue RNA-seq - ScienceDirect
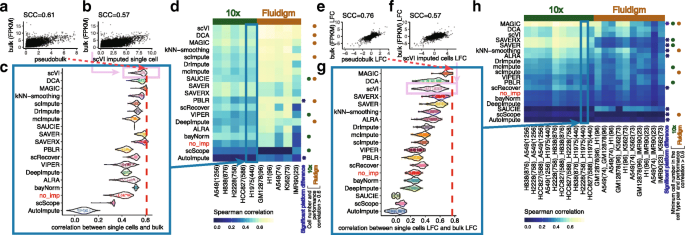
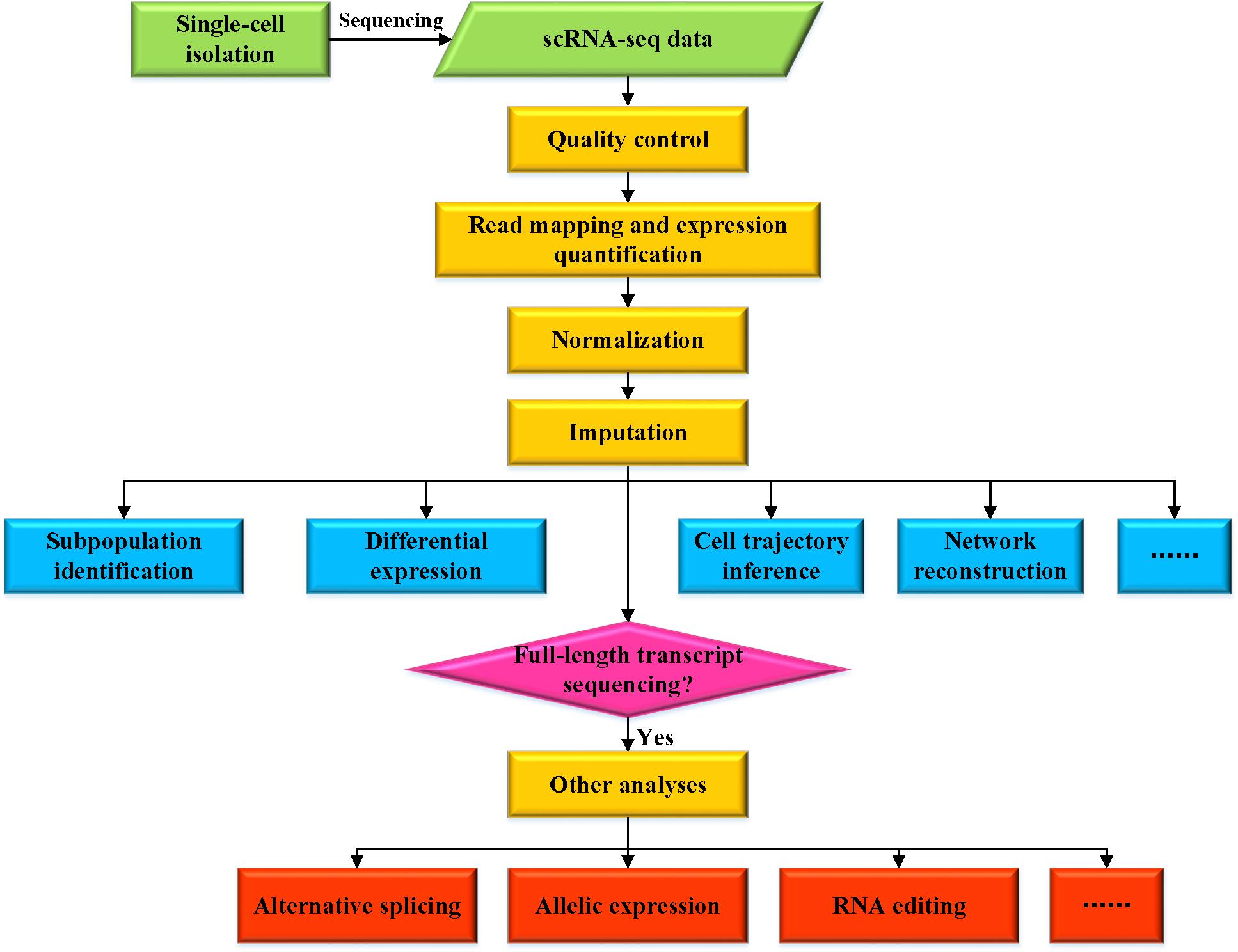
A systematic evaluation of single-cell RNA-sequencing imputation methods | Genome Biology | Full Text
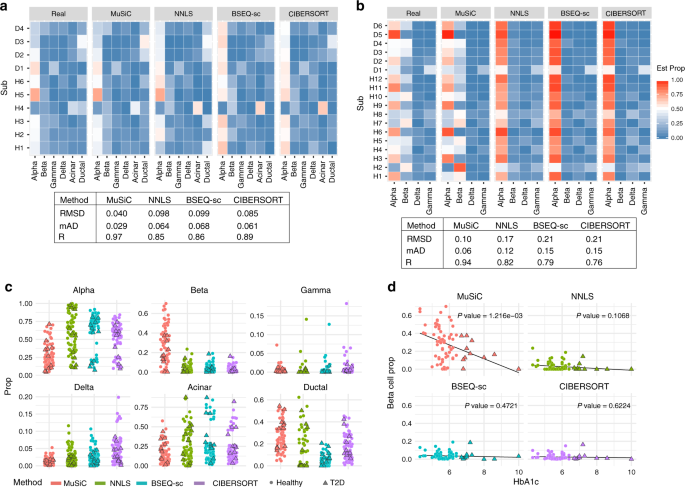
Bulk tissue cell type deconvolution with multi-subject single-cell expression reference | Nature Communications
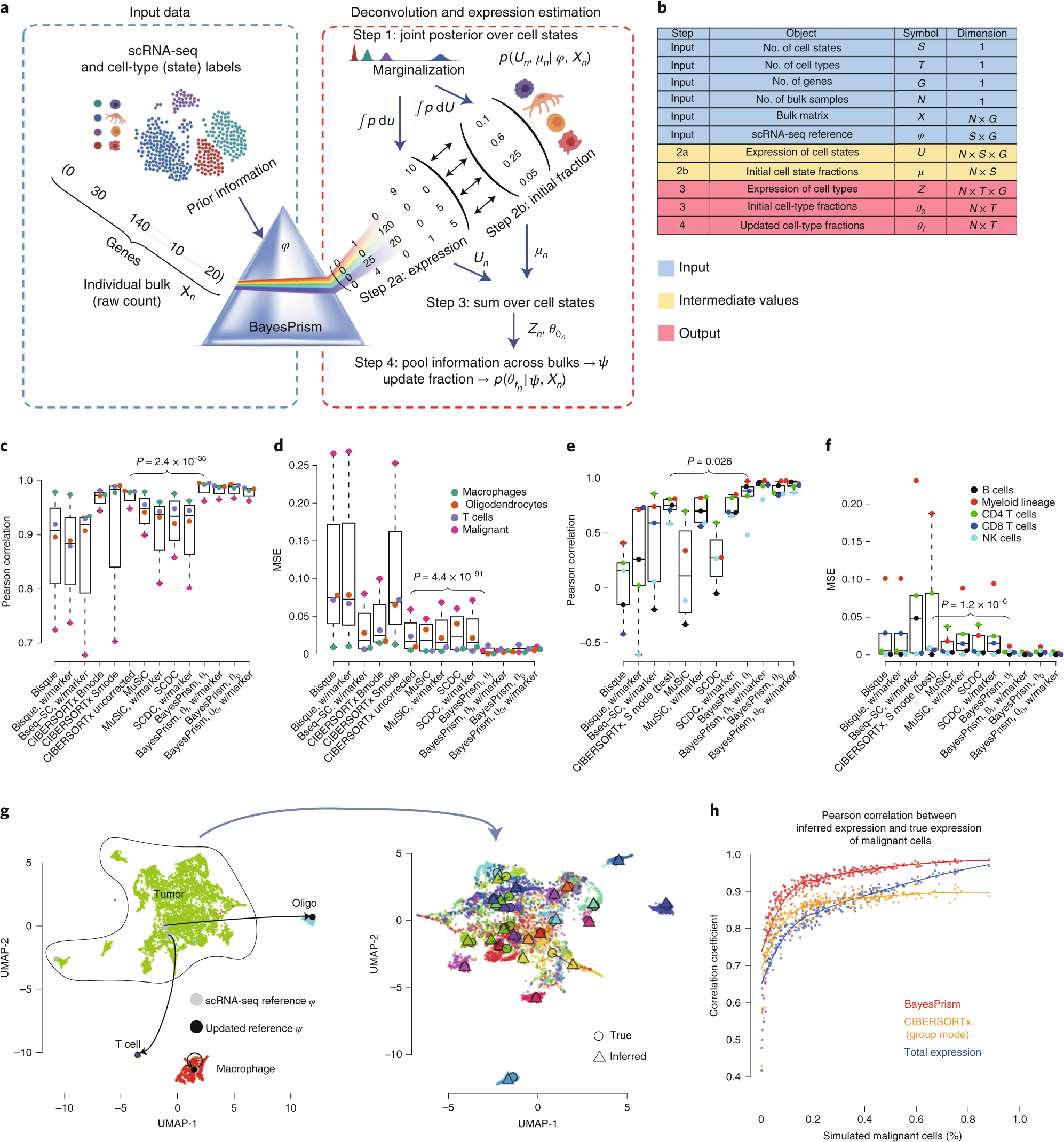
Cell type and gene expression deconvolution with BayesPrism enables Bayesian integrative analysis across bulk and single-cell RNA sequencing in oncology | Nature Cancer

SCDC – bulk gene expression deconvolution by multiple single-cell RNA sequencing references | RNA-Seq Blog

Cellular deconvolution of GTEx tissues powers discovery of disease and cell-type associated regulatory variants | Nature Communications
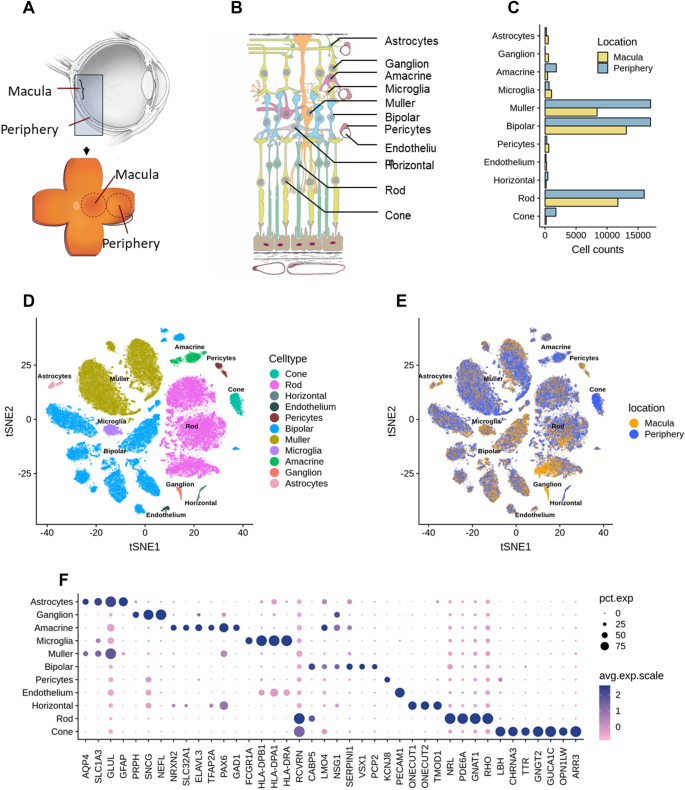
Implication of specific retinal cell-type involvement and gene expression changes in AMD progression using integrative analysis of single-cell and bulk RNA-seq profiling | Scientific Reports
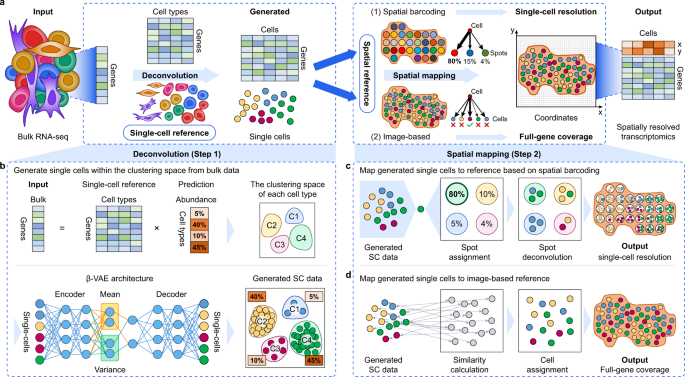
De novo analysis of bulk RNA-seq data at spatially resolved single-cell resolution | Nature Communications

A protocol to extract cell-type-specific signatures from differentially expressed genes in bulk-tissue RNA-seq: STAR Protocols